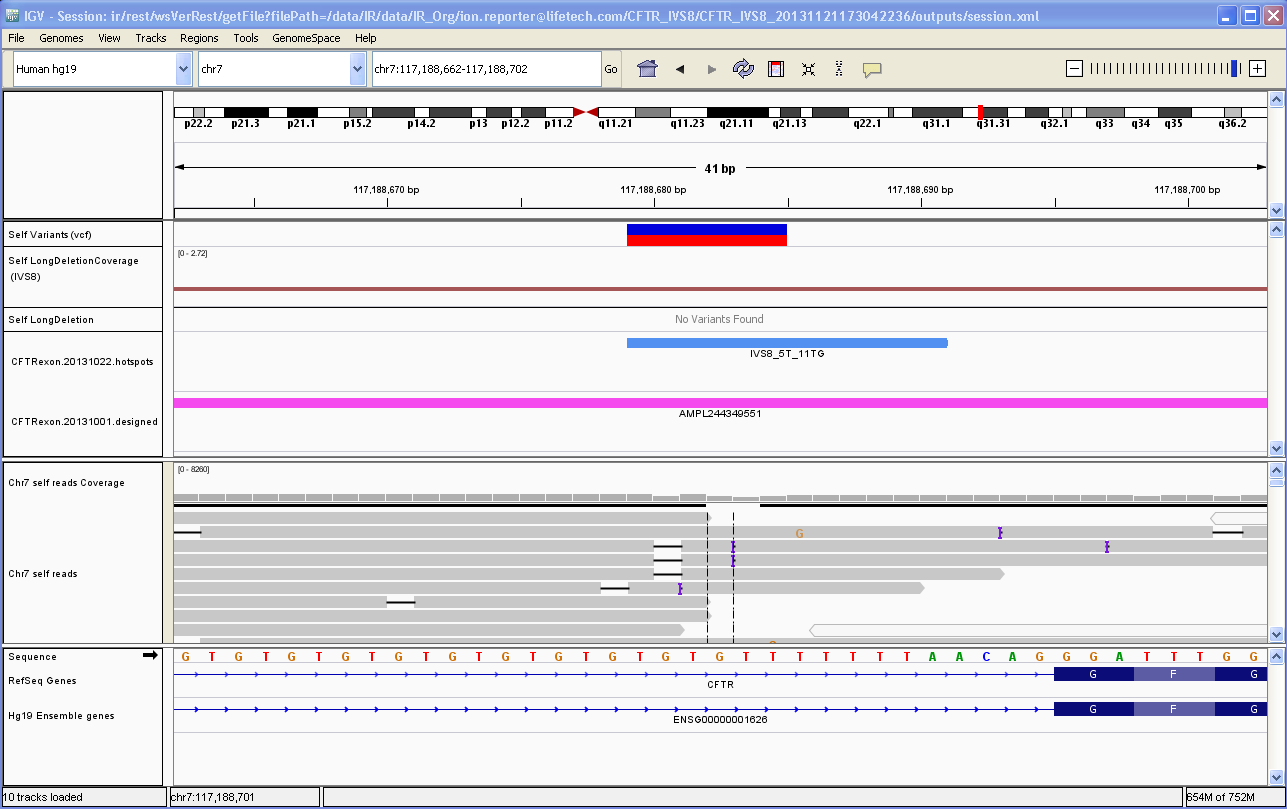
Visualize variants with IGV
You can visualize data from Ion Reporter™ Software with the Broad Institute Integrative Genomics Viewer (IGV). The viewer is available at the Broad Institute website: http://software.broadinstitute.org/software/igv/home.
If you visualize different analyses in IGV that use the same samples or the same panel files (but different algorithm versions, for example), the reads, read counts, BED file, and other tracks load into the browser only one time. In this case, if the software detects exact duplicate tracks, all unique tracks load separately if the software detects any differences in results data or input tracks between multiple analyses for visualizations of the requested analyses.
You must have IGV set as the default viewer in Ion Reporter™ Software before you start this procedure. For more information, see Set IRGV or IGV as the default viewer.
The following steps are for a Chrome™ browser on the Windows™ operating system. The instructions vary slightly based on the operating system and browser that you use.
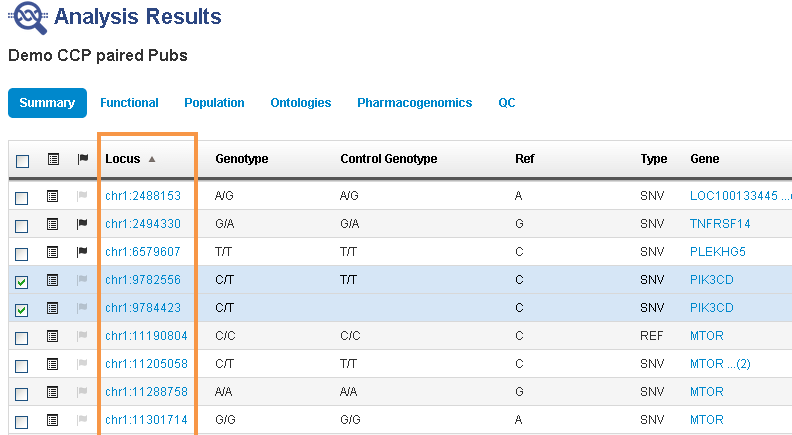
- In the Analysis tab, click the name for the analysis of interest to open analysis results.
-
In the Analysis Results screen, click the locus for a variant of interest.
- In the notice that appears in the browser download bar, click Keep.
- Do one of the following steps to open an IGV visualization for a variant: