Data types for gene fusions analyses
Use the following guidelines to interpret the fusion calls and other information that is includes in analysis results. In addition to fusion type assays, the analysis results and visualization include information on assays of other types:
-
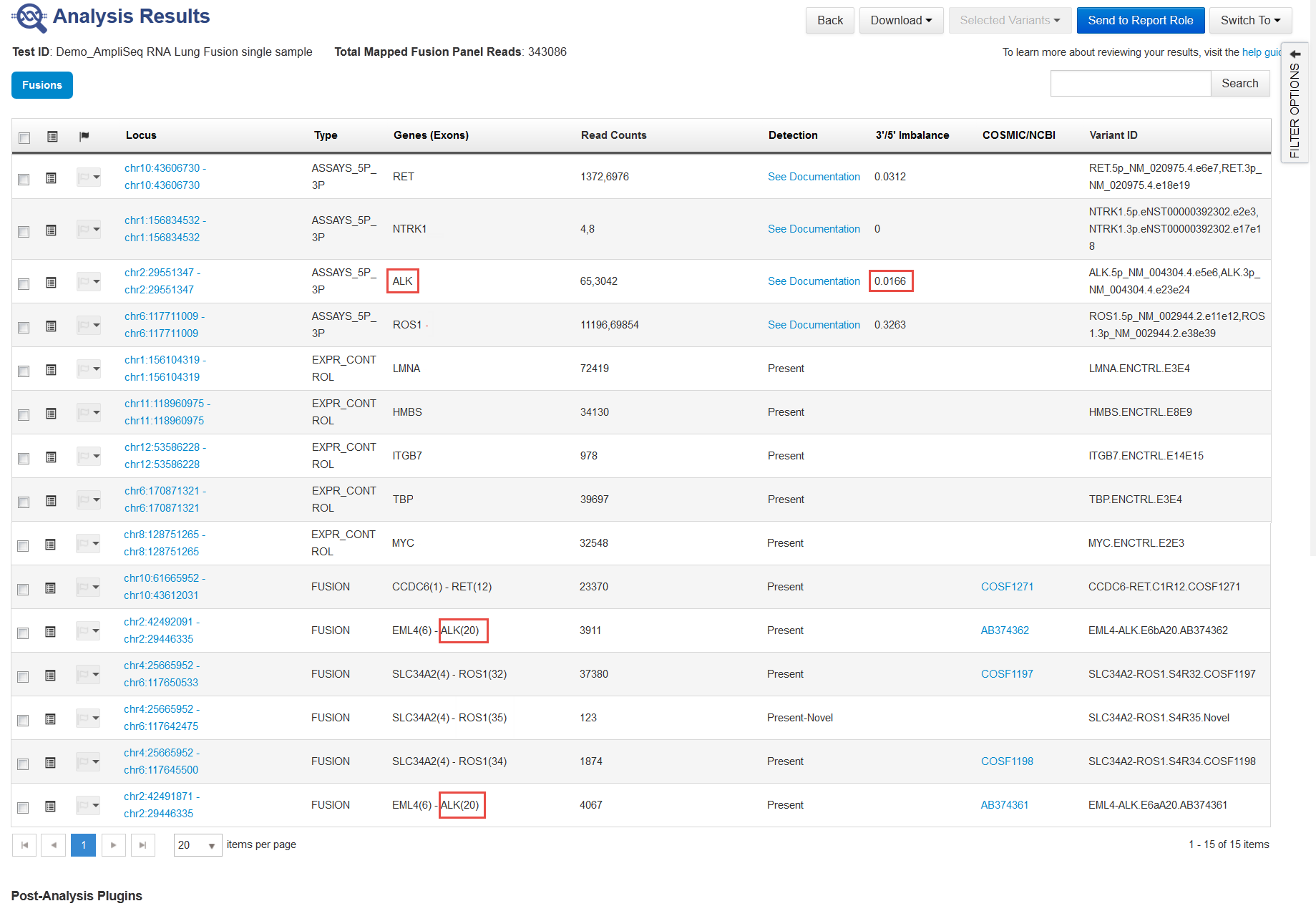
ASSAYS_5P_3P—5' and 3' assays provide confirmation for the fusion calls. This score is reported in the 3'/5' Imbalance column.
Four assays are built into the Ion AmpliSeq™ RNA fusion panels. Each assay corresponds to one of the acceptor genes in the panel, and that corresponding acceptor gene is reported in the Genes (Exons) column.
For example, the assay reported in the third column names ALK in the Genes (Exons) column. For example, in rows 10 through 15 in this example, the 3'/5' Imbalance score for this assay reflects the strength of the calls for all fusions with the ALK driver gene.
The Genes (Exons) column does not report exon numbers for assays rows.
-
EXPR_CONTROL—Few expression control assays are built into the panel to confirm the success of the sequencing run that the analysis is based on.
A limited number of expression control genes are built into the Ion AmpliSeq™ panel for quality checking purposes. If multiple EXPR_CONTROL calls are reported as Absent in the Detection column, check the Torrent Suite™ Software run report for the sequencing run, as this result could indicate a low-quality run or failed amplification.
-
Fusion— Fusion assays are built into the panels to confirm the presence of the targeted fusion isoforms and non-targeted fusion isoforms in a sample. For more information, see Fusion detection methods and Fusion calls in analysis results.
-
ProcessControl—A few control assays are built into the TagSeq and Ion AmpliSeq™ HD panels to help determine if the sample is valid or not, similar to the Expression control assays
-
GENE_EXPRESSION—These assays provide confirmation for the Gene expression amplicons that are built into the panel and their level of expression.
-
RNA_HOTSPOT—These assays provide confirmation for SNPs or INDELs at a particular position (hot spot).
-
RNAExonVariant—These assays provide measurement of expression for intragenic events such as Exon Deletions, Exon skipping events, Alternate Splice transcripts and Wild type transcripts. For these assays, two additional metrics are reported.
-
-
For all the RNAExonVariant assays that belong to a Gene with at-least one RNAExonVariant type assay. It is designed to amplify the wild type transcript of that gene and report this value.
-
-
For all the RNAExonVariant type assays, this normalized count is reported.
Norm count within Gene for RNAExonVariant Vi = (Read Count of Vi) / (Sum of read counts of all RNAExonVariants of the same Gene).
Detection column for RNAExonVariant will have " N/A " as the value for all the RNAExonVariants.
-
-
RNAExonTile—RNAExonTile assays are designed across a few exon-exon junctions in each fusion driver gene. The measurement of expressions from these assays per each gene can be used to confirm the presence of a fusion in the sample. For more information, see Fusion detection methods.
Note: You can view the NORM_COUNT_TO_HK metric in the variants VCF file. The normalized expression to housekeeping value is the read count of a target normalized by the average expression of housekeeping genes rather than by the total number of mapped reads. The value is calculated from the read count: