Searches on the Analysis Results screen
Advanced searches, such as those that use OR and AND, can be performed on the Analysis Results screen with a controlled vocabulary query language. The searches are available in both a single-analysis variant review table and a multi-sample visualization table. However, the searches have been tested only on the single-analysis variant review table.
Note: Most searches on the Analysis Results screen can be done effectively with filters, instead of with the search field. For most searches, use a filter instead.
The following filter terms are supported:
|
Notation |
Meaning |
|---|---|
|
key:value1,value2 |
key=value1 OR key=value2 |
|
key:[min,max] |
min <=key <=max |
|
key:(min,max) |
min < key < max |
|
key:[min,] |
min <= key |
|
key:[,max] |
key <= max |
|
key:(min,) |
min < key |
|
key:(,max) |
key < max |
|
key:value* |
key contains value |
|
key:* |
key exists (key has any value) |
|
-key:value |
key != value |
The following keys are supported:
|
Key |
Example |
|---|---|
|
locus |
chr2:123456 |
|
function |
missense |
|
location |
exonic |
|
cosmic, omim, pfam, drugbank, go |
glioma |
|
dbsnp |
rs12345 |
|
gene, transcript |
TP53, NM_01010.1 |
|
maf |
[0.0,0.05] |
|
coverage |
[1000'] |
|
sift, polyphen, grantham |
['0.05] |
|
type |
INDEL |
|
comment |
something* |
|
vkb "vkb" stands for MyVariants. |
* |
Search field behavior is different on the variant table in the Analysis Results screen, than for searches on other screens.
Asterisks are useful to search for matches in any annotation source. (By contrast, a filter search matches only one annotation source.) Asterisks (*) for some variant table searches on the Analysis Results screen is used in the following ways:
-
An asterisk (*) in the search field is allowed only on the Analysis Results screen.
-
An asterisk (*) is required for some searches, but is not allowed for other searches. The differences are due to how the different types of information are stored.
-
The asterisk is a search wildcard. Without the asterisk, searches match only the exact string entered. With asterisks both before and after the search string, matches at the beginning, middle, and end are all found.
-
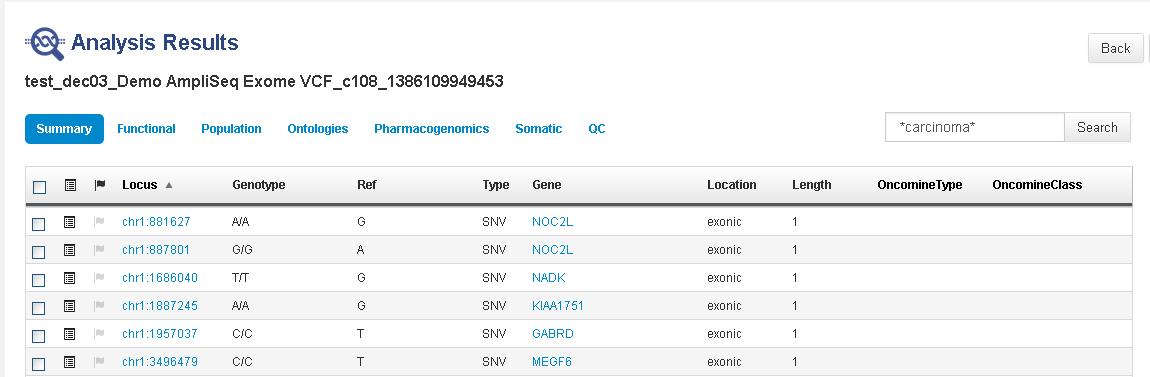
This example shows a search for *carcinoma*.
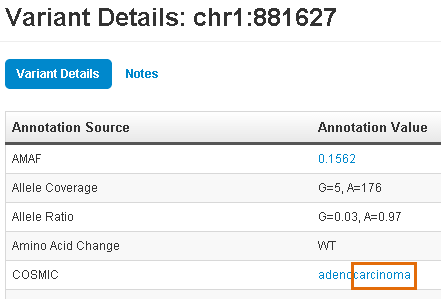
This search lists all variants that have an OMIM, COSMIC, ClinVar, DrugBank, and so on, annotation that contains "carcinoma" anywhere in the annotation. Click
(Details) for one
(Flag) of the matched variants to open its variant detail card. Scroll down to find the entries that contain "carcinoma".
-
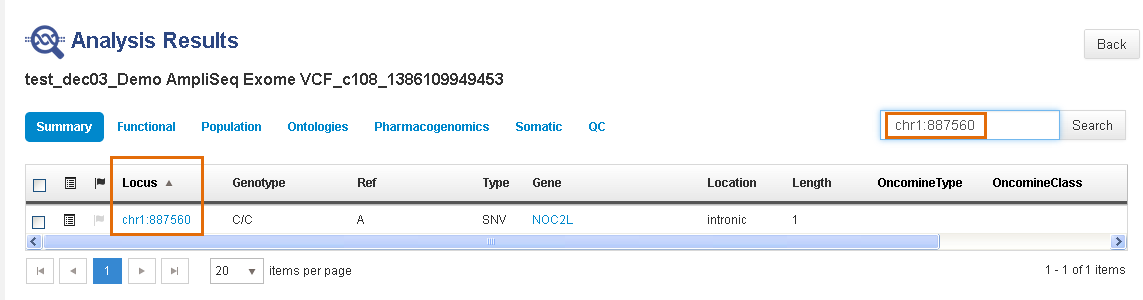
To search for a locus, enter the complete locus entry, with chromosome number and full position number.
-
Do not use an asterisk (*) for a locus search. Searches on a chromosome number by itself or with a partial position number also are not supported.
-
For other information, use a filter.
-
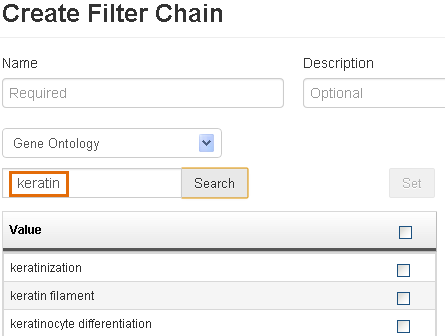
Asterisks cannot be used in search terms during creation of a filter chain.
-
(gene:TP53 OR (function:missense,nonsense AND (maf:[0.0,0.05} OR -dbsnp:*))) OR type:CNV
This retrieves all variants that fall in TP53, all CNVs, plus variants that have a functional impact of missense or nonsense AND either have a minor allele frequency less than 0.05 or are novel (not found in dbSNP).
-
cosmic:carcinoma* AND ((type:SNV AND sift:[0.0,0.10]) AND coverage:[300,] OR locus:chrX
This retrieves all SNV variants annotated with COSMIC histology terms containing "carcinoma" with a deleterious SIFT score ( <0.10) and high coverage(>300), plus any variant that falls on chromosome X.